
The reference alignment, which includes the traditional 27f and 1492r
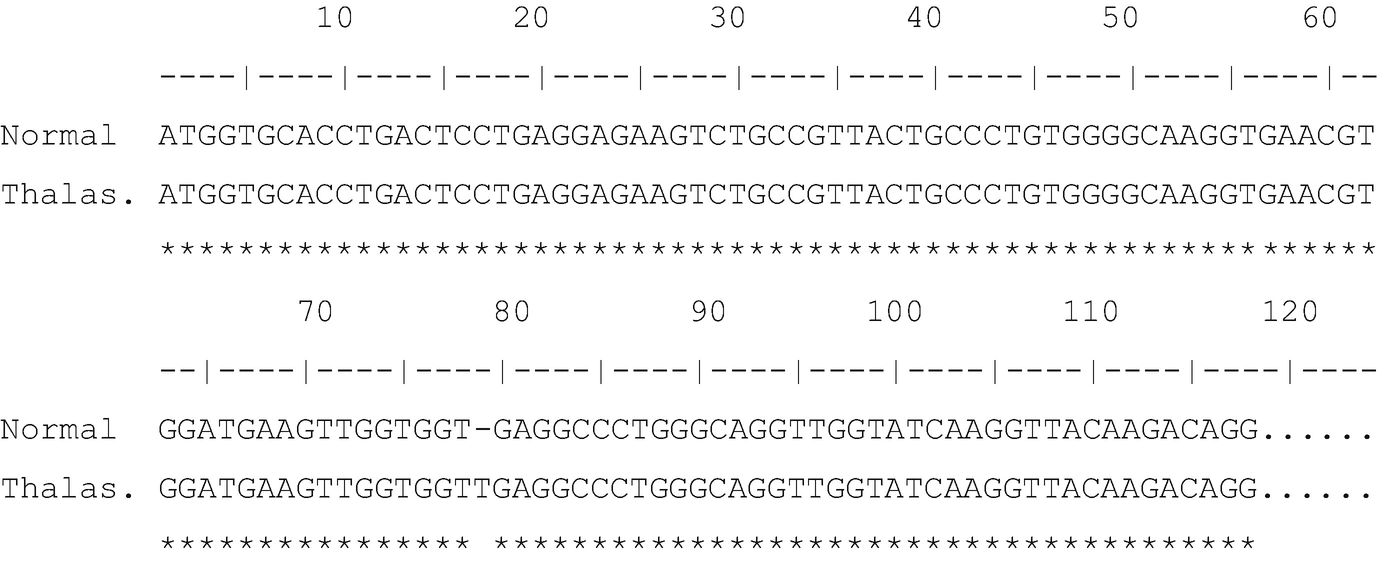
This occurred because the sequenceĪctually has vector sequence at the 5’ end, which is not represented in In this example we see that the alignment actually starts atĬandidate sequence position 5. Method, which resulted in a pairwise alignment that was 501 characters Next, we see that we used the needleman alignment
In this example we see that candidate sequence AY457915 was 501 bases QueryName QueryLength TemplateName TemplateLength SearchMethod SearchScore AlignmentMethod QueryStart QueryEnd TemplateStart TemplateEnd PairwiseAlignmentLength GapsInQuery GapsInTemplate LongestInsert SimBtwnQuery&TemplateĪY45793 1525 kmer 89.07 needleman 5 501 1 499 499 2 0 0 97.60 To run the aligner enter the following command:
We will use the greengenes alignment with 7,682 Located in the same folder as your candidate file and where you are The qs command requires the user to enter a candidate and Various features of this command, we will use the AbRecovery Least be implicitly based on the secondary structure. Secondary structure of the 16S rRNA gene, if the template database isīased on the secondary structure, then the resulting alignment will at While the aligner doesn’t explicitly take into account the Furthermore, this rate can be accelerated using multiple The SILVA alignment in less than 3 hrs with a quality as good as the

Very fast - we are able to align over 186,000 full-length sequences to Silva alignments however, custom alignments forĪny DNA sequence can be used as a template and users are encouraged to Gotoh, or blastn algorithms and iii) to re-insert gaps to the candidateĪnd template pairwise alignments using the NAST algorithm so that theĬandidate sequence alignment is compatible with the original template Suffix tree searching ii) to make a pairwise alignment between theĬandidate and de-gapped template sequences using the Needleman-Wunsch, The closest template for each candidate using kmer searching, blastn, or The qs command aligns a user-suppliedįasta-formatted candidate sequence file to a user-suppliedįasta-formatted template alignment.


 0 kommentar(er)
0 kommentar(er)
